import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.datasets import make_classification
#from sklearn.tree import DecisionTreeClassifier, export_graphviz
#from sklearn.model_selection import train_test_split
#from sklearn.preprocessing import OneHotEncoder
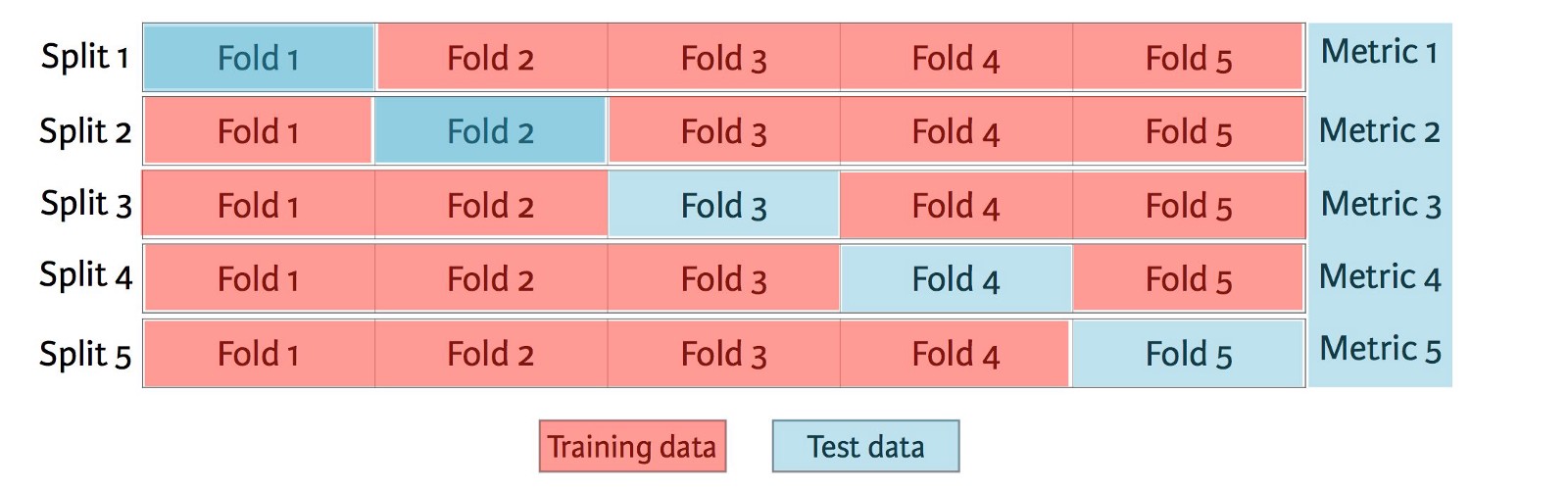
K-Fold Cross Validation¶
K-Fold cross validation is a validation technique in which the training data is split into K (roughly) evenly sized portions, called folds. K versions of a model are contructed using the same hyperparameters. Each model is trained on K-1 folds and validated on the remaining fold. Each fold is used as a validation set for exactly one model. The overall validation performance of the model is reported as the average of the K scores.
Common values for K are 3, 5, 10, and n-1. When K = n-1, we refer to the technique as "Leave-one-out cross-validation".
We will see several ways of implemention cross-validation in this lesson.
Generate Data¶
np.random.seed(1)
X, y = make_classification(n_samples=250, n_features=6, n_informative=6, n_redundant=0, n_classes=7, class_sep=2)
multipliers = [0.01, 100, 0.1, 10, 1, 5]
for i in range(0,6):
X[:,i] = multipliers[i] * X[:,i]
np.set_printoptions(suppress=True, precision=2)
print('Distribution of Features:')
print('Min: ', np.min(X, axis=0))
print('Max: ', np.max(X, axis=0))
print('Mean:', np.mean(X, axis=0))
print('SDev:', np.std(X, axis=0))
np.set_printoptions(suppress=True, precision=4)
Naive Implementation of 5-Fold Cross-Validation¶
tr_acc = []
va_acc = []
for k in range(0,5):
mask = np.ones(250).astype('bool')
mask[50*k : 50*(k+1)] = False
temp_mod = LogisticRegression(solver='lbfgs', multi_class='ovr', max_iter=1000)
temp_mod.fit(X[mask,:], y[mask])
tr_acc.append(temp_mod.score(X[mask,:], y[mask]))
va_acc.append(temp_mod.score(X[~mask,:], y[~mask]))
print('Training Scores: ', tr_acc)
print('Validation Scores:', va_acc)
print('Avg Valid Score: ', np.mean(va_acc))
Note that the validation scores vary somewhat considerably across the different folds. If we were to perform traditional validation, we would have very different impressions of the performance of our model if the validation set was equal to the 2nd fold as opposed to the 5th fold.
Using cross_val_score¶
Scikit-Learn comes with several tools for performing cross validation. The most basic of these is cross_val_score.
from sklearn.model_selection import cross_val_score
mod_01 = LogisticRegression(solver='lbfgs', multi_class='ovr', max_iter=1000)
cv_results = cross_val_score(mod_01, X, y, cv=5, scoring='accuracy')
print(cv_results)
print(np.mean(cv_results))
models = [
LogisticRegression(C=1, solver='lbfgs', multi_class='ovr', max_iter=1000),
LogisticRegression(C=10, solver='lbfgs', multi_class='ovr', max_iter=1000),
KNeighborsClassifier(n_neighbors=5),
KNeighborsClassifier(n_neighbors=10),
DecisionTreeClassifier(max_depth=2, random_state=1),
DecisionTreeClassifier(max_depth=4, random_state=1),
SVC(kernel='rbf', C=1, gamma=0.1),
SVC(kernel='rbf', C=1, gamma=1)
]
cv_scores = []
for mod in models:
cv_results = cross_val_score(mod, X, y, cv=5, scoring='accuracy')
cv_scores.append(np.mean(cv_results))
print(np.array(cv_scores))
idx = np.argmax(cv_scores)
print(models[idx])
Using KFold for Cross-Validation¶
If we need more flexibility when performing cross-validation, we can use the KFold class from sklearn. This class can be used to create the folds that we will use, but it does not fit any models. This is useful when, for instance, we need to preprocess our data.
from sklearn.model_selection import KFold
five_fold = KFold(n_splits=5, shuffle=True, random_state=1)
split = five_fold.split(X, y)
for train_index, val_index in split:
print(val_index)
from sklearn.preprocessing import StandardScaler
five_fold = KFold(n_splits=5, shuffle=True, random_state=1)
cv_scores = []
for train_index, val_index in five_fold.split(X, y):
X_train, y_train = X[train_index, :], y[train_index]
X_val, y_val = X[val_index, :], y[val_index]
scaler = StandardScaler()
Xs_train = scaler.fit_transform(X_train)
Xs_val = scaler.transform(X_val)
fold_scores = []
for mod in models:
mod.fit(Xs_train, y_train)
fold_scores.append(mod.score(Xs_val, y_val))
cv_scores.append(fold_scores)
cv_scores = np.array(cv_scores)
cv_means = np.mean(cv_scores, axis=0)
print(cv_scores)
print()
print(cv_means)
idx = np.argmax(cv_means)
print(models[idx])
After identifying the preferred model using cross validation, you should retrain that model on the entire training set.
svm_mod = SVC(kernel='rbf', C=1, gamma=1)
svm_mod.fit(X, y)
print('Training Accuracy:', svm_mod.score(X,y))
Using StratifiedKFold for Cross Validation¶
The StratifiedKFold class from sklearn behaves similarly to KFold, except that it attempts to create folds in which the distribution of the classes in the individual folds are similar to that of the original set.
from sklearn.model_selection import StratifiedKFold
strat_five_fold = StratifiedKFold(n_splits=5, random_state=1)
strat_cv_scores = []
for train_index, val_index in strat_five_fold.split(X, y):
X_train, y_train = X[train_index, :], y[train_index]
X_val, y_val = X[val_index, :], y[val_index]
scaler = StandardScaler()
Xs_train = scaler.fit_transform(X_train)
Xs_val = scaler.transform(X_val)
fold_scores = []
for mod in models:
mod.fit(Xs_train, y_train)
fold_scores.append(mod.score(Xs_val, y_val))
strat_cv_scores.append(fold_scores)
strat_cv_scores = np.array(strat_cv_scores)
strat_cv_means = np.mean(strat_cv_scores, axis=0)
print(strat_cv_scores)
print()
print(strat_cv_means)
Comparison of KFold and StratifiedKFold Results¶
print('--Mean of CV Scores--')
print('Ordinary: ', cv_means)
print('Stratified:', strat_cv_means)
print()
print('--StdDev of CV Scores--')
print('Ordinary: ', np.std(cv_scores, axis=0))
print('Stratified:', np.std(strat_cv_scores, axis=0))
Leave-One-Out Cross Validation¶
We can perform leave-one-out cross validation by using KFold with the number of splits equal to the number of observations.
loo = KFold(n_splits=250, shuffle=False, random_state=1)
loo_scores = []
for train_index, val_index in loo.split(X, y):
X_train, y_train = X[train_index, :], y[train_index]
X_val, y_val = X[val_index, :], y[val_index]
scaler = StandardScaler()
Xs_train = scaler.fit_transform(X_train)
Xs_val = scaler.transform(X_val)
fold_scores = []
for mod in models:
mod.fit(Xs_train, y_train)
fold_scores.append(mod.score(Xs_val, y_val))
loo_scores.append(fold_scores)
loo_scores = np.array(loo_scores)
loo_means = np.mean(loo_scores, axis=0)
print(loo_means)
Scikit-learn also has a LeaveOneOut class for performing leave-one-out cross validation.
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
loo_scores = []
for train_index, val_index in loo.split(X, y):
X_train, y_train = X[train_index, :], y[train_index]
X_val, y_val = X[val_index, :], y[val_index]
scaler = StandardScaler()
Xs_train = scaler.fit_transform(X_train)
Xs_val = scaler.transform(X_val)
fold_scores = []
for mod in models:
mod.fit(Xs_train, y_train)
fold_scores.append(mod.score(Xs_val, y_val))
loo_scores.append(fold_scores)
loo_scores = np.array(loo_scores)
loo_means = np.mean(loo_scores, axis=0)
print(loo_means)